feature
Dogma Spies
Cracking the Code to Teaching Mutation
Modeling is an important tool in science teaching and learning. Constructing a model instead of replicating one is more meaningful and better supports student learning than analyzing premade models (Firooznia 2015; Gouvea and Passmore 2017; Takemura and Kurabayashi 2014). Models include physical replications of a scientific phenomenon or analyzing premade diagrams, both of which will have limitations and should be addressed with students. Modeling processes that students cannot physically see can help them make more meaningful connections than analyzing diagrams of the processes (Firooznia 2015; Reinagel and Bray Speth 2016). Specifically with central dogma (the overall process of reading DNA to create proteins), students can experience a disconnect between how DNA mutations can affect proteins, as well as how mutations can impact organisms and evolution (Prevost, Smith, and Knight 2016; Zhao and Schuchardt 2019). Modeling mutations can help students focus on what is most relevant during the process (Reinagel and Bray Speth 2016). This mutations lesson for high school students involves analyzing how point mutations occur and affect genetic codes, while also engaging in developing and using models, one of the science and engineering practices of the NGSS.
Central dogma
Central dogma is often considered an abstract and complicated process that is challenging for students (Firooznia 2015; Prevost, Smith, and Knight 2016; Reinagel and Bray Speth 2016; Takemura and Kurabayashi 2014). Transcription involves reading DNA and creating a complementary strand of messenger RNA (mRNA), replacing the nitrogenous base thymine in DNA with uracil in RNA. Students can have trouble matching proper nitrogenous bases with each other, which is necessary for more complex genetic information (Newman et al. 2018). Translation occurs when a ribosome “reads” the RNA in bases of three, called codons. Each codon matches a specific amino acid brought to the ribosome by transfer RNA (tRNA). If the amino acid matches the mRNA codon, it and other amino acids will form a polypeptide chain (Takemura and Kurabayashi 2014). For example, if the codon is GUU, the amino acid it matches is valine. Students often understand the general flow of DNA to RNA to protein but struggle with what truly occurs during the processes of transcription and translation (Briggs et al. 2016).
Lessons on central dogma often culminate with mutations. While rare in eukaryotic organisms, mutations can alter genetic sequences, which result in changes in RNA and possibly leading to a different protein (Prevost, Smith, and Knight 2016). Mutations are necessary for genetic variation, a concept touched upon in several NGSS standards and integral to genetics and evolution. It is important for students to explore mutations and evaluate the effects of mutations. Point mutations modify the individual bases in DNA. Frameshift mutations occur when a base is added (an insertion) or removed (a deletion), shifting the genetic code and subsequently the amino acid order of a protein. A missense mutation happens when a base is changed, causing the amino acid to change. Nonsense mutations can be the most detrimental to protein synthesis, where a base change causes a STOP codon to replace an amino acid, often shortening the RNA strand (Prevost, Smith, and Knight 2016). Silent mutations alter the DNA sequence, but the same amino acid is added to the polypeptide chain, making no difference to overall protein production (Zhao and Schuchardt 2019).
This two-day lesson should occur after students have already learned transcription and translation. This 9th-grade lesson reinforces the process of protein synthesis with students, as well as allows them to create their own mutations and analyze the effects. Allowing students to construct their own models allows them to see what is most relevant when mutations form and provides a physical representation of the effects on proteins (Newman et al. 2018; Reinagel and Bray Speth 2016). Presented through the exciting lens of a spy story, students act as secret agents to address NGSS HS-LS3-2. This lesson also incorporates asking questions, developing models, and constructing explanations, which are NGSS science and engineering practices. If faced with the challenge of hybrid teaching, this lesson can be altered to involve online students. When the lesson was taught in a hybrid setting, the students were told ahead of time what supplies would be needed in case they had them at home. Otherwise, the online students worked as scribes and guides to the students in the classroom.
Day 1
Engage
To begin Day 1, play spy-themed music as the students walk in. Place the students in groups of two to three people. Explain to the students they have been recruited by the CIA to work as secret agents specializing in breaking codes. Guide the students to the CIA Spy Kids website, where under the Games tab they select “Break the Code” to reach the activity (see Online Connections), then either allow them to choose a code to break or assign a code to break that is relevant to this lesson. Allow the students five minutes to decode the message, then the following questions can be asked:
- What did you do in this activity?
- How did you decide on what to do to break the code?
- What did you have to look for or use to decipher the code?
- What connections can you make between spy codes and mutations?
- What would happen to the code if one letter was changed? One letter removed or added?
- What do you wonder about changes in proteins?
If there is time and the students do not organically connect spy codes with mutations, play a short YouTube video titled, “The Genetic Code Animation” (see Online Connections). This video also provides a clear review of translation as well as a smooth transition to the Explore portion of this lesson.
Explore
Place the students in groups of two to three again or allow them to continue to work with their previous group. When this lesson was conducted in a classroom, the students stayed in the same groups as the Engage portion; however, this is flexible for teachers to fit their classroom needs. Once together, assign each group of the following point mutations:
- Deletion
- Insertion
- Frameshift
- Silent
- Nonsense
Students should be given 15 minutes to construct a presentation to teach the other groups about their point mutation. The presentation should include the following: the mutation, how it affects DNA, an example of how the DNA mutation affects the RNA strand and amino acid chain, and a real-world mutation of its kind. For example, the deletion group could present on how a deletion happens when a base is removed from the genetic code, changing the sequence and subsequently the RNA strand. This would then alter the amino acid chain because the codons would all be different. The real-world effect of a mutation could be cystic fibrosis (Figure 1). Learn.Genetics.Utah.edu has an interactive page where students can explore mutations such as extra toes in cats and curly hair in dogs (Learn.Genetics), then they can add one to their presentation.
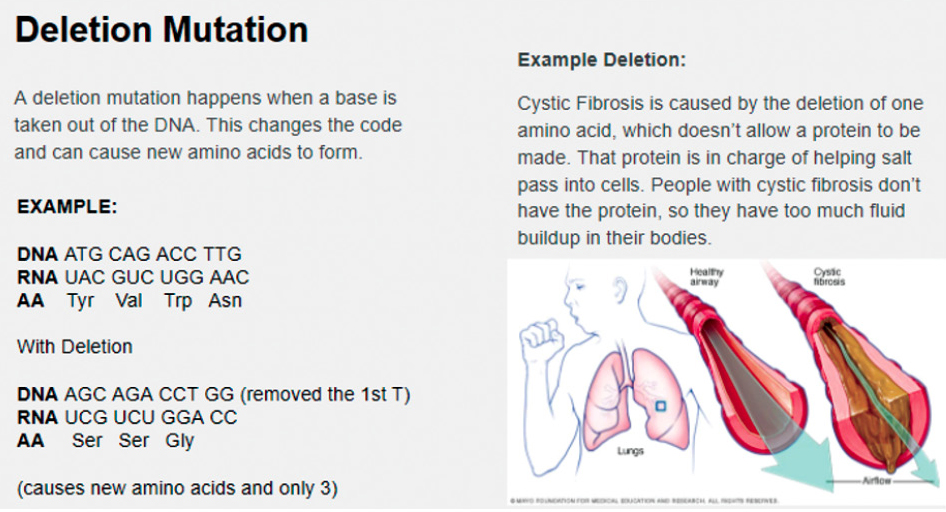
Cystic Fibrosis deletion mutation.
This portion of the lesson can be modified depending on technology, class culture, and class size. The students can search the internet on their own or websites can be provided. The National Human Genome Research Institute (Genetics Glossary). is a great resource, with an online glossary of genetic terms in English and Spanish. If in a hybrid teaching situation, have at least one student on campus in a breakout room with online students. This allows the students to collaborate and is easier to manage. Encourage the students to be creative and visual. Avoid providing examples or definitions; allow time for the students to synthesize what their mutation is on their own.
Explain
End the first day’s activities with each group taking two minutes to present their mutations to the class. As the students present, offer feedback and ask each group a question tailored to the mutation (Table 1).
This is the time to correct misunderstandings and reaffirm themes—such as where the mutation occurred and what type of effect it had on the result of a protein—and to gauge students’ evaluation of the impact the mutation would have on a protein. This can be done by asking probing questions that prompt students to use evidence from the activity to explain their claim.
At this point, students may have trouble connecting how the mutations affect the protein once translation has occurred. Have an example DNA strand ready for the students to guide you through the process of transcribing and translating it. Once the amino acid chain has been determined, ask students where a mutation could occur. Then they can choose one to mutate the DNA strand (they can choose any point mutation except a silent mutation). The example included here (Figure 2) is an insertion. As a class, go back and transcribe and then translate the mutated DNA strand to analyze the effects of the chosen mutation. If there is not time to complete this portion of the lesson, it can be a class warm-up activity on Day 2. Rock Pocket Mice and Predation on the HHMI BioInteractive website would also be good homework for review during Day 2 (see Online Connections).
| Table 1. Potential questions to ask during the presentations. | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
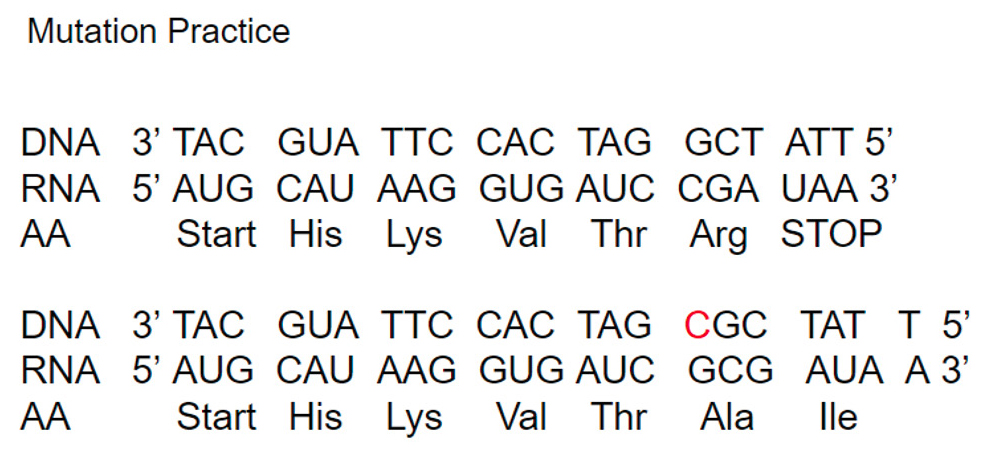
Example of Insertion Mutation.
Day 2
Elaborate
At the beginning of Day 2, students have a chance to create their own secret code! This secret agent–themed laboratory activity prompts students to create an RNA strand bracelet based on a provided DNA strand. Different bead colors code for different amino acids. For 20–30 minutes, students work in pairs to mutate the DNA strand using whatever point mutation they want. For example, after the students have created their bracelet, they may choose to delete one of the bases from their DNA. They would transcribe the mutated DNA into RNA, then translate the RNA into amino acids. The students will see how the bead colors change from their original bracelet. This will be done twice to provide students a chance to practice two mutations. The students will then remake the code bracelet for a visualization of the changes. The codes can be delivered in envelopes for extra security and the students can play the role of “agents.”
The following materials are needed for this laboratory activity (Figure 3):

Code bracelet materials.
- One DNA secret code and mission assignment per group (six different codes provided in the teacher resources)
- One pipe cleaner per student
- 11 plastic cups labeled with amino acid names
- 11 different colors of pony beads (about 25 beads per color)
- One amino acid chart per pair of students
Separate the beads by color, then place them in labeled cups for each amino acid. The cups can be spread out around the classroom for students to collect. As the Spymaster, pass out the secret code and Mission Assignment to each group (add a “Top Secret” envelope for extra flair!), as well as a pipe cleaner for every student. The students can then proceed through the laboratory activity at their own pace while the teacher circulates. If you are teaching in a hybrid environment, pair the online students with students in the classroom to help build the bracelets.
Once the students construct their “mission bracelets,” the teacher can use them as a fast way to formatively assess the students’ mastery of protein synthesis (Figure 4). If approved to move forward with the mission, the students will scramble the code twice using two different point mutations of their choice. Afterward, the students can go around the room and recollect their beads to redo the bracelet and see how the mutations affected it. If the students create any amino acid not included in the list, they will use a purple bead. After the students finalize their bracelets, it is up to the teacher whether the students can keep them or return the materials.
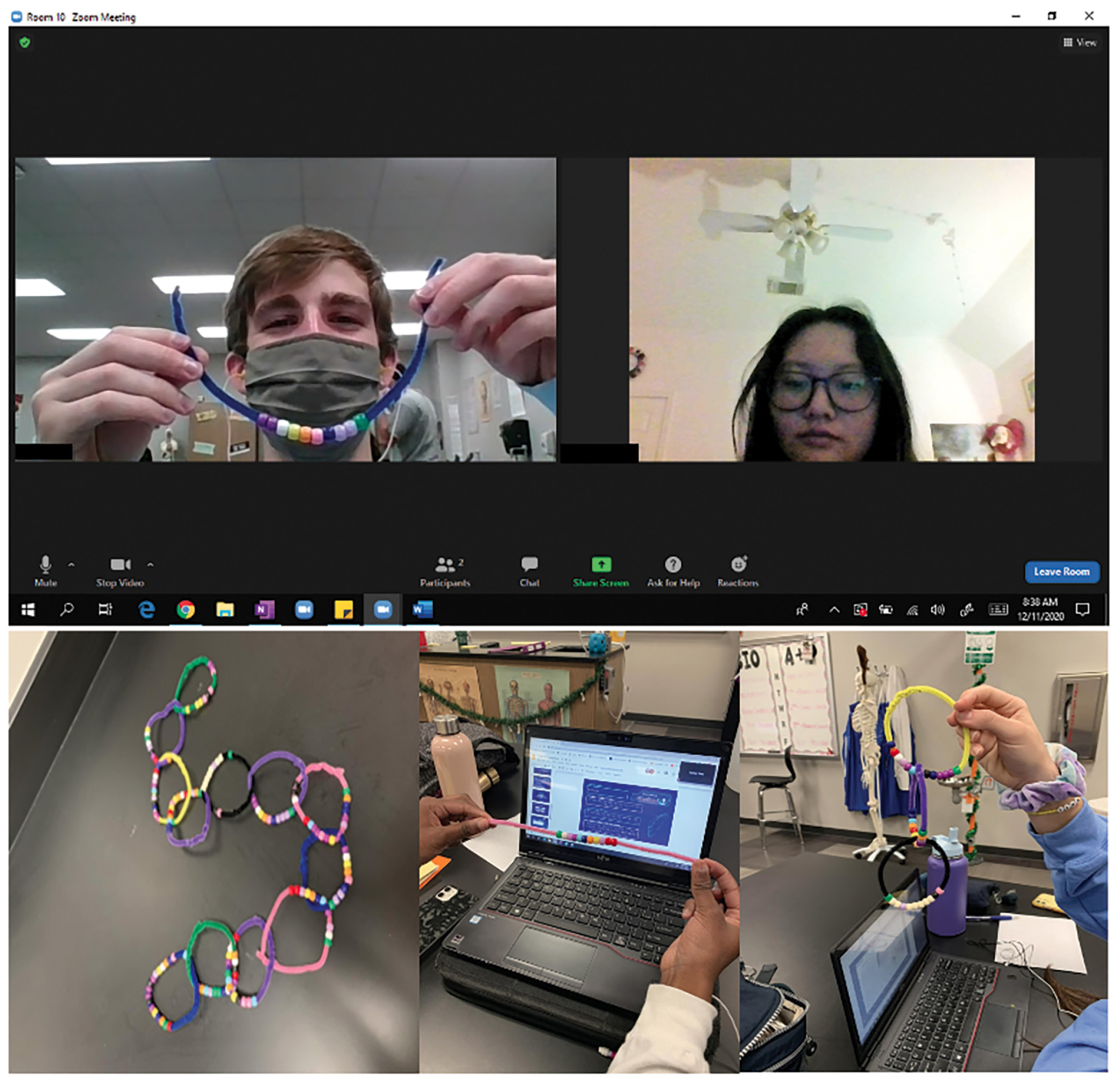
Images of student generated code bracelets.
Evaluate
At the end of the lab, there is a 10- to 20-minute long “Mission Debrief” in which students evaluate their bracelets and mutations, as well as reflect on their learning (Figure 5). Additionally, an exit ticket will assess students’ understanding about whether mutations are always harmful; students should defend their reasoning. Students can explain how mutations contribute to genetic diversity in terms of evolutionary change. The students also analyze two DNA strands and determine the type of mutation that occurred. The exit ticket includes an additional challenge question to evaluate the students’ ability to transcribe and translate RNA with mutations. The exit ticket could be done during class or can be sent home as homework.

Examples of student responses to mission debrief.
Conclusion
This lesson provides a great way for teachers to quickly assess a student’s understanding of transcription and translation from previous lessons. It also provides students ownership of the mutations and allows them to creatively scramble their codes. Common misunderstandings to check for while facilitating include the 5’ and 3’ designations on the RNA strands and whether the students separate their RNA into codons. The students should also be checked for accuracy on how point mutations affect the strands. This lab can easily be scaled down or up, depending on classroom needs. For higher-level learners, they could make their initial DNA sequences on their own instead of receiving one from the teacher. There are also resources on the BioInteractive website applicable for advanced students for further exploration of mutations (HHMI BioInteractive). The lesson can also be chunked and guided depending on student needs. Whatever the learning levels of students, this lesson can help them construct models of what could be too abstract to visualize on their own. Adding the beads to the bracelets for each amino acid, then changing them provides students visual and kinesthetic associations with how mutations alter DNA and subsequently protein synthesis. As mutations are a major contributor to genetic variation, students who have a solid understanding of how they happen can better appreciate evolution. ■
Online Connections
CIA Spy Kids: https://www.cia.gov/spy-kids/games/index.html
The Genetic Code Animation: https://www.youtube.com/watch?v=Q9WlnEpyvpY
National Human Genome Research Institute genetics glossary) https://www.genome.gov/genetics-glossary
HHMI Biointeractive website: https://www.biointeractive.org/
Student exit ticket: https://bit.ly/3bjGlcw
Teacher resources: https://bit.ly/39HiiE8
Connecting to the Next Generation Science Standards: https://bit.ly/3zYdipz
Cassandra Montalto is a doctoral student in the Department of Curriculum and Instruction at the University of Houston. Sissy S. Wong is an Associate Professor of Science Education in the Department of Curriculum and Instruction at the University of Houston.
5E Biology Curriculum Evolution Labs Lesson Plans NGSS STEM Teaching Strategies